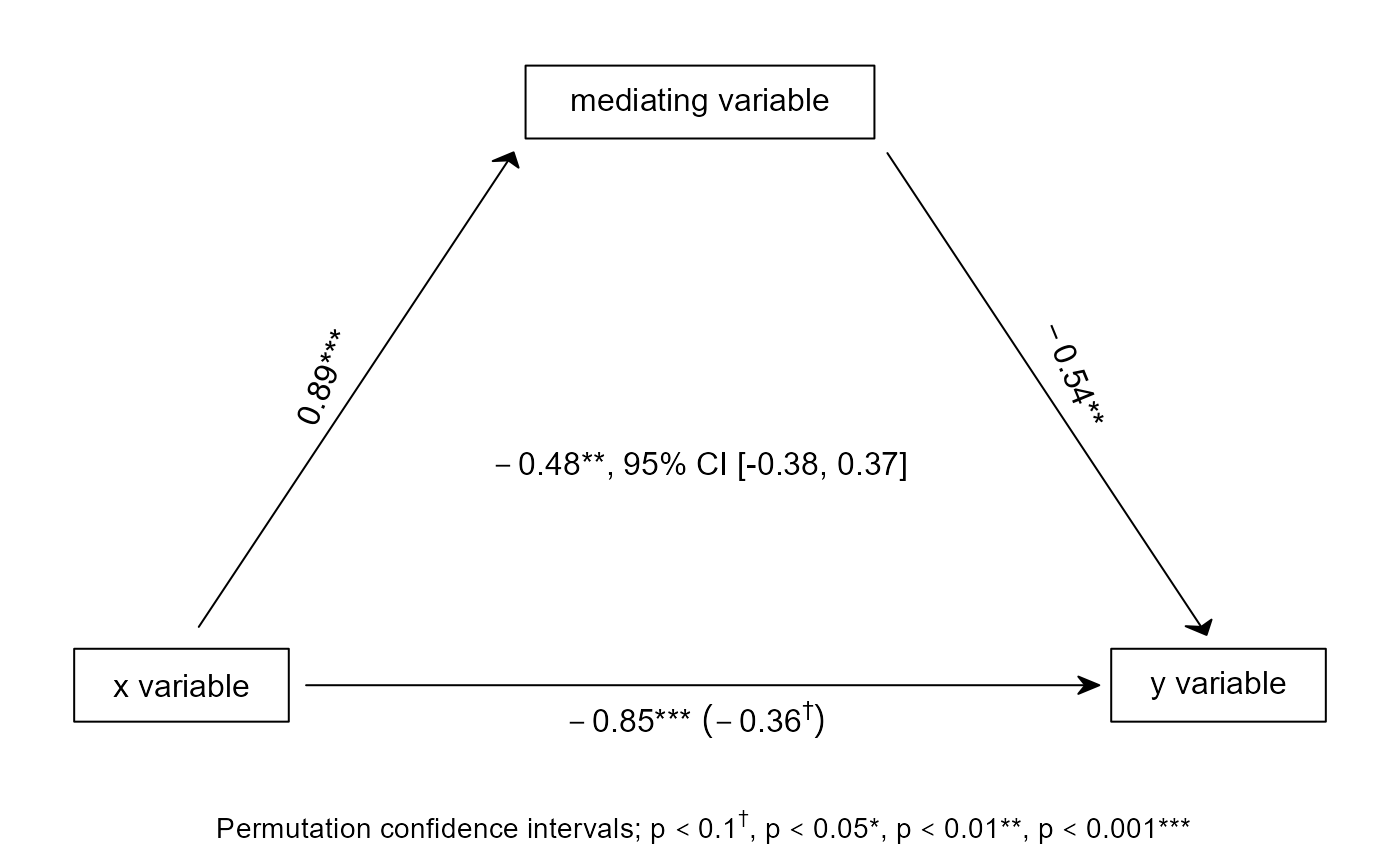
Plot simple mediation figures, maybe displaying indirect effects and confidence intervals of some sort, and potentially saving them.
Usage
medfig(mat, ci = NULL, ci.type = NULL, xlab = "x variable",
ylab = "y variable", mlab = "mediating variable", title = FALSE,
note = TRUE, box = TRUE, digits = 3, note.sig = TRUE,
fin.length = 0.5, cords.box = list(), cords.arrow = list(),
padding.box = 0.05, padding.arrow = 0.02, padding.number = 0.02,
padding.title = 0.1, padding.note = 0.1, arrow.scale = 0.01,
box.fill = NA, center.adj = 0.1, color = c(line = NA, box = NA, label =
NA, number = NA, title = NA, note = NA), weight = c(line = NA, box = NA,
label = NA, number = NA, title = 2, note = 0.9), save = FALSE,
format = pdf, name = "mediation", ...)Arguments
- mat
Output from
bspmed, or a matrix with two rows and 4-5 columns (see example).- ci
A vector of confidence intervals (one lower and one upper). If the entries are named, these will be read as numbers and added together for the displayed percent (see example).
- ci.type
Text to be displayed in the note (i.e.,
ci.typeconfidence intervals).- xlab, ylab, mlab
Text to be displayed in each variable box.
- title, note, box
Logical indicating whether to draw each element.
- digits
Number of digits to round to.
- note.sig
Logical indicating whether asterisks corresponding to p-values should be added to numbers.
- fin.length
Length of the arrowhead fins, back from the line intersection (0 would be a flat base, for a triangular arrowhead).
- cords.box
Center position of each box. Should be a list with at least one coordinate pair (a vector with values for y and x position). If entries aren't named, these x, y, and m are assigned as names; coordinates pairs can be entered positionally or named. Default is
list(x = c(.1, .1), m = c(.5, .9), y = c(.9, .1)).- cords.arrow
Position of each arrow. This behaves in the same way as cords.box, but entries are names a, b, and c, and each set of coordinates should be 4 values specifying start and end positions. Any unspecified coordinates are calculated based on box positions.
- padding.box, padding.arrow, padding.number, padding.title, padding.note
Specifies the padding around each of the elements; box adjust space around texts in each box, arrow adjust distance from boxes, and number adjusts space between numbers and lines.
- arrow.scale
Size of each arrowhead.
- box.fill
Color of the box backgrounds.
- center.adj
Amount to shift the center text downward (positive values making center text closer to the bottom).
- color
A vector with named entries corresponding to any of the elements (line, box, label, number, title, note). Box refers to the color of the line (whereas
box.fillspecifies the background). Single unnamed values are applied to all elements.- weight
The weight of arrow lines or box outlines, or size of numbers or texts. Behaves the same as color.
- save, format, name
Information used when saving the figure to a file; if any are specified, an image is saved. Format refers to the graphics engine; default is
pdf. Some elements may not show up properly using other engines (such ascairo_pdf). Other engines are included in the baselink[grDevices]{grDevices}package. Another option isemffrom thedevEMFpackage (used to create the example image).- ...
Additional arguments passed to par (parameters of the plot frame).
See also
bspmed to run a mediation model.
Examples
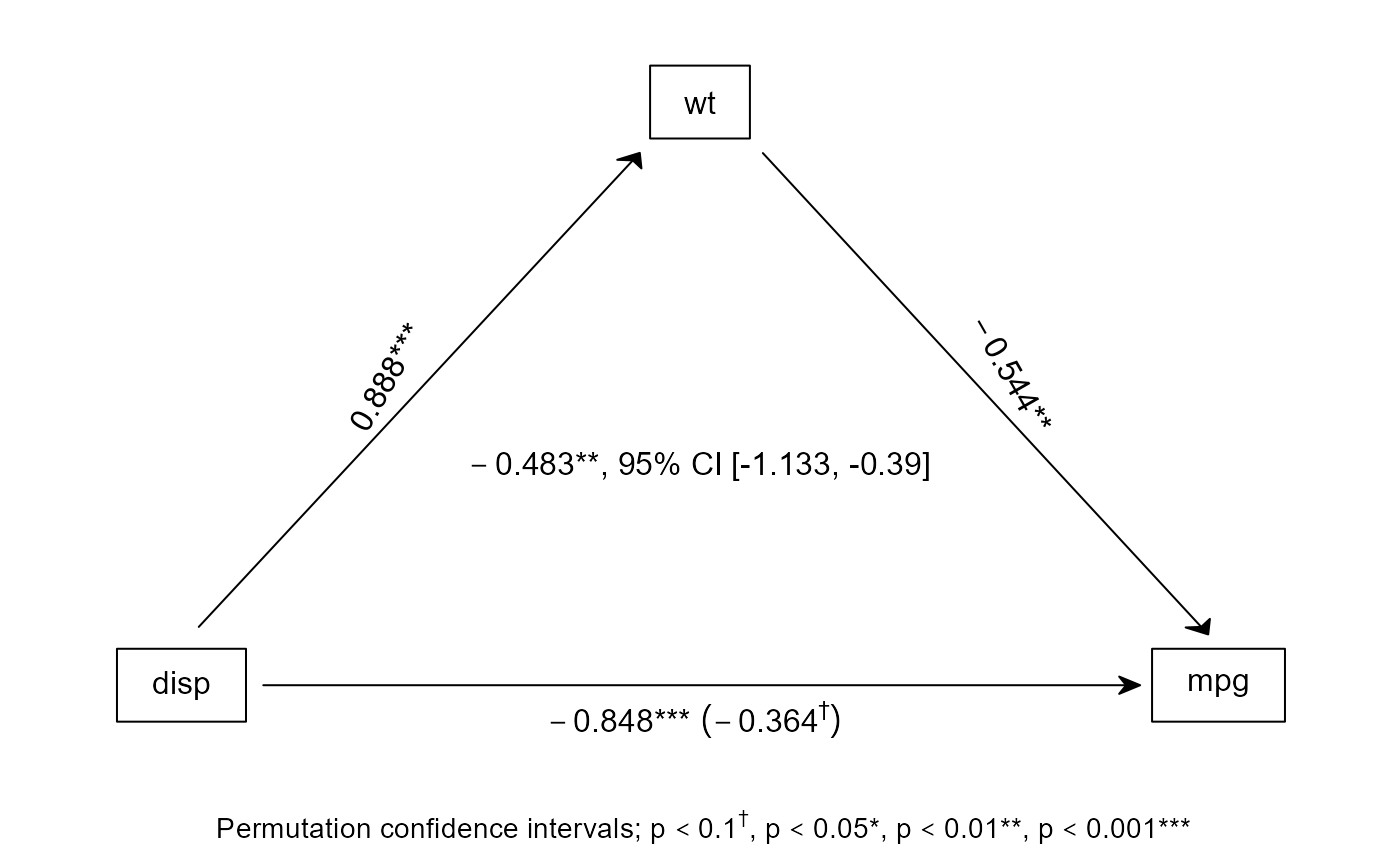
# by default, the bspmed function will show a figure
bspmed(disp, mpg, wt, scale(mtcars))
#> Effect of disp on mpg, by way of wt
#> Processed in 0.99 seconds
#>
#> Sobel
#> a b c c' indirect
#> b 0.888 -0.5440 -0.8476 -0.3645 -0.4831
#> p 0.000 0.0074 0.0000 0.0636 0.0055
#>
#> Permutation confidence intervals
#> 2.5% 97.5% all <|> -0.4831
#> CI -1.132512 -0.3899162 FALSE
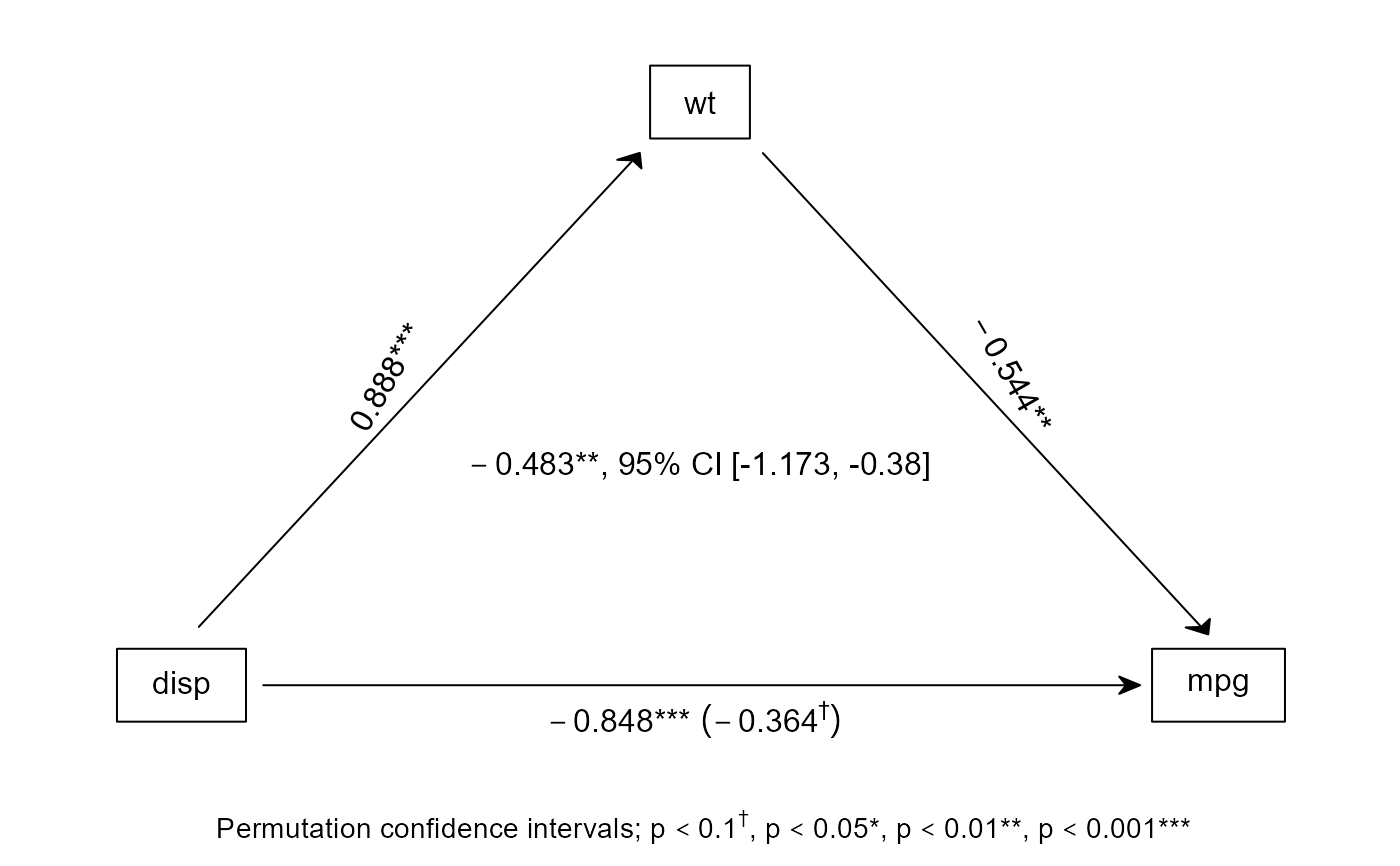
 # you can also save the output from a bspmed call and plot a figure for it later:
med <- bspmed(disp, mpg, wt, scale(mtcars), figure = FALSE)
#> Effect of disp on mpg, by way of wt
#> Processed in 0.99 seconds
#>
#> Sobel
#> a b c c' indirect
#> b 0.888 -0.5440 -0.8476 -0.3645 -0.4831
#> p 0.000 0.0074 0.0000 0.0636 0.0055
#>
#> Permutation confidence intervals
#> 2.5% 97.5% all <|> -0.4831
#> CI -1.173451 -0.3800334 FALSE
medfig(med)
# you can also save the output from a bspmed call and plot a figure for it later:
med <- bspmed(disp, mpg, wt, scale(mtcars), figure = FALSE)
#> Effect of disp on mpg, by way of wt
#> Processed in 0.99 seconds
#>
#> Sobel
#> a b c c' indirect
#> b 0.888 -0.5440 -0.8476 -0.3645 -0.4831
#> p 0.000 0.0074 0.0000 0.0636 0.0055
#>
#> Permutation confidence intervals
#> 2.5% 97.5% all <|> -0.4831
#> CI -1.173451 -0.3800334 FALSE
medfig(med)
 # Or you can enter a matrix, with confidence intervals in the second position,
# and what type they are in the third.
mat <- matrix(
c(.89, .0001, -.54, .001, -.85, .0001, -.36, .05, -.48, .001),
2,
dimnames = list(c("b", "p"), c("a", "b", "c", "c'", "i"))
)
medfig(mat, c("2.5" = -.38, "97.5" = .37), "Permutation")
# Or you can enter a matrix, with confidence intervals in the second position,
# and what type they are in the third.
mat <- matrix(
c(.89, .0001, -.54, .001, -.85, .0001, -.36, .05, -.48, .001),
2,
dimnames = list(c("b", "p"), c("a", "b", "c", "c'", "i"))
)
medfig(mat, c("2.5" = -.38, "97.5" = .37), "Permutation")